The DNA molecule is relatively unstable compared with other cellular components and will degrade with time if not repaired. As soon as an organism dies, its cells rupture releasing enzymes which rapidly digest strands of DNA; nutrient-rich fluids, which encourage the growth of environmental microorganisms, contribute to further degradation. However, under certain conditions, such as rapid desiccation or burial in anaerobic conditions, freezing, low soil pH, or high salt concentrations, decay enzymes and microorganisms are inactivated before DNA is completely disassembled. In these cases, DNA can remain preserved in short fragments of 100-500 base pairs for remarkably long periods.
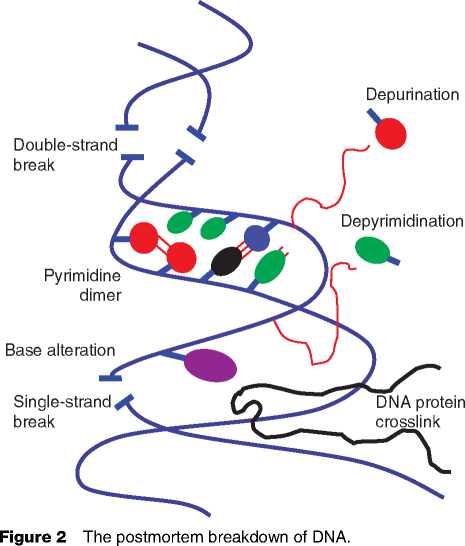
These short fragments of DNA are also subject to chemical damage by radiation, oxidation, condensation, and hydrolysis (Figure 2). Hydrolysis breaks the backbone of DNA strands while oxidation blocks polymerase action, and condensation cross-links proteins and DNA. These forms of damage can stall or confuse the enzymes used in PCR to give erroneous results; for example, the most common form of damage (the deamination of cytosine residues) causes a characteristic nucleotide change during sequencing (a C-to-Tor G-to-A change).
The upshot of these processes is that aDNA specimens frequently contain very little of original DNA, which is why most aDNA studies focus on mitochondrial rather than nuclear DNA: there are simply more copies of mitochondria, so the chances of getting an aDNA sequence are that much higher.
(b)
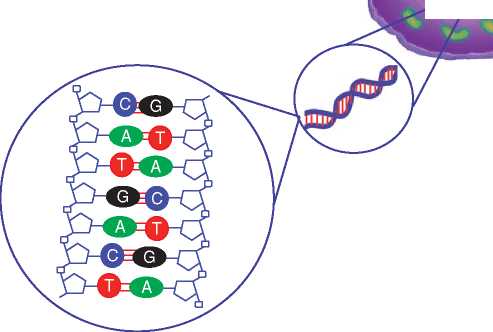
Figure 1 Cracking the jargon. (a) A cell has one nucleus and many mitochondria. (b) DNA is made up of four bases: adenine, thymine, guanine and cytosine which bind together to make a double stranded helix.
(a)

 World History
World History









