There are a great number of methods for the analysis of aDNA, and new methods are being developed continuously. Therefore, this is a snapshot of commonly used methods available at the time of publication.
In the Extraction Lab:
Clean samples. Using bleach, buffered solutions, and UV light, particulate matter (e. g., soil) and extraneous DNA from living organisms are removed.
Mobilize DNA. DNA is brought into solution to be separated from other components of the cell and organism. It usually involves grinding the sample into a fine powder, which is mixed for several hours in an extraction buffer, containing a digestive enzyme (Figure 3(a)).
Purify DNA. This removes all but the DNA from the buffer including the remaining particles of the sample (e. g., bone powder, crushed seed, etc.) and any other remaining proteins, sugars, soil particles, etc. that have also come into solution with the DNA. There are many available methods for this step.
Precipitate/concentrate DNA. This ensures that you have a high enough concentration of DNA to be able to make the amplification step work. Sometimes, other steps may be added to enhance results; for example, the use of DNA repair enzymes or glycosyl bond-cleaving agents.
In the PCR set-up room (where even the air is filtered and UV irradiated):
Prepare for DNA amplification. A mixture of template aDNA, a buffer, magnesium salt, and a pair of Primers designed to target a specific section of DNA is made up. This is then taken to the postextraction lab where PCR and downstream processing is carried out.
In the PostExtraction Lab:
PCR
PCR works like a photocopier and makes multiple copies of the DNA you ask it to target. PCR can be difficult from ancient material for a number of reasons (inhibition by co-extracted compounds, very low copy number of DNA, DNA swamped by contaminants, DNA too badly fragmented or damaged), but there are a number of methods to help improve success rates.
Visualize amplified DNA. Different lengths of DNA are separated out on an electrophoresis gel. DNA is stained with ethidium bromide and fluoresces under UV light and can easily be scanned by eye for a DNA fragment of the expected size (Figure 3(b)).
Read DNA sequence. DNA can now be sent for sequencing (Figure 3(c)). This ‘direct sequence’ is a consensus of the pool of amplified DNA, which may include errors from damaged DNA and possibly even contaminant sequences. However, to understand how damaged and mixed your sample is you must use bacterial cloning.
Bacterial cloning. A single fragment of DNA resulting from PCR (i. e., a single photocopy) is inserted in a bacterial host, which then copies it many thousands of times. Then the DNA from multiple bacterial colonies are sequenced separately and the sequences are compared to look for error and contamination. Finally, a consensus sequence is constructed from these multiple sequences and checked for statistical robustness.
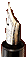

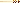
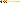
 World History
World History









